Conn Lab @ The University of Pennsylvania
Overview - RNA translation for select gene expression in disease
Insights into the genetic landscape of patient samples have depicted an explosion of diagnostic and clinical biomarkers, yet not all changes at the transcription level translate into proteomic expression. Genetic alterations also remain difficult to target and many oncogenes play roles for normal cellular functions which prevents their targeting. Despite these limitations, there arise phenotypic vulnerabilities that malignant cells rely on for their survival and the dynamic regulation of RNA helps to orchestrate these cascades.
KEYWORDS & TECHNIQUES: Cancer Biology, Adaptive Stress Responses, mRNA Translation, epitranscriptomics - RNA modifications, Drug targeting, Protein Homeostasis, small molecule drug screening, Ribosome Profiling, Proteomics, in vivo and ex vivo disease modeling
Post-transcriptional gene regulation for homeostasis & cell survival
DNA is targeted for transcription, RNA spliced, processed, & exported into the cytoplasm…
and yet without selection for protein synthesis gene expression would be halted.
. . .
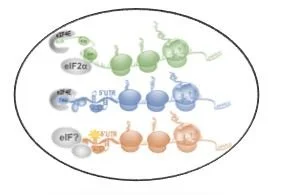
The dynamic response of RNA translation allows cells to adapt gene expression for survival. These steps are orchestrated by RNA modifications, RNA binding proteins, and the signaling cascades that impinge on them. We focus on the interplay of these processes.
Cancer initiation & progression
Drivers of cancer initiation have documented the complexity and synergy that exist during tumorigenesis. Understanding why one cell progresses into aggressive disease and another corrects through cell death, remains poorly understood. The heterogeneity of cancer cell populations requires further investigation to access aggressive disease earlier and target accurately.
Adaptive stress responses & noncanonical gene expression
We discovered that adaptive stress responses can activate select post-transcriptional regulatory pathways that directly promote cancer progression & metastasis.
. . . These pathways can translate unique protein isoforms, release long or short peptides, and therefore expand the genetic repertoire of a cancer cell. Identifying the regulators of these processes and characterizing the novel functions of isoforms is a major goal in the lab.
Goals:
Identifying the signaling pathways that are activated in cancer cells allowing for their dynamic adaptations to stress (nutrient, proteomic, oxidative...etc) during cellular transformation and disease progression
Determine how aggressive cancer cells rewire their translational machinery to select distinct RNAs for peptide production giving ‘life’ to our genes, while…
Characterizing unique isoforms created through non-conventional initiation and the functions they serve
Most importantly, elucidating the relevance of the above processes towards disease detection, improved therapeutic targeting, and treatments with an underlying goal for prevention.
Our research has been made possible with support from these organizations:
Collaborations:
Past:
Peter Walter- UCSF
Ralph DeBerardinis- UT Southwestern
Shingo Kajimura- Harvard Medical School
David Fruman- UC Irvine
Amy Lee- University of Southern California
Ongoing:
Baillis Lab- CHOP
Weitzman Lab- CHOP
Jonathan Edwards- CHOP, PSOM
Fan Lab- PSOM
Koumenis Lab- PSOM
Kelly Jordan-Sciutto & Judy Grinspan- PSOM
Conine Lab- CHOP